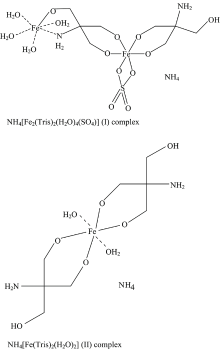
The reaction of iron(Ⅲ) anions, FeCl3 and Fe2(SO4)3 with tris(hydroxymethyl) aminomethane (Tris) in a 1:2 molar ratio affords the new coordination compounds NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ) and NH4[Fe(Tris)2(H2O)2] (Ⅱ). These compounds were characterized by elemental analysis, and their molecular structures were determined by spectroscopic methods (infrared and electronic spectra), magnetic susceptibility, and molar conductivity measurements, and further corroborated by thermo gravimetric analysis and its differential (TGA/DrTGA). According to the experimental data, the complexes can be characterized in the solid state as mono- and binuclear, with a distorted octahedral stereochemistry. The distorted octahedral stereochemistry adopted by the complexes was confirmed by the magnetic susceptibility measurement of NH4[Fe2(Tris)2(H2O)4(SO4)], which consists of a six-coordinate iron atoms in a distorted octahedral environment constructed from four O atoms (two Tris molecules), two O atoms from the loosely associated SO4 coordinated ligand, and O, N of second Tris molecule with occupying by four oxygen atoms of coordinated water molecules. Regarding, NH4[Fe(Tris)2(H2O)2] complex the mono iron atom is surrounded by six oxygen atoms as four by two Tris molecules and two coordinated water molecules in axial form. Antibacterial and anticancer activities of the complexes were studied and the complexes were screened against bacteria, colorectal adenocarcinoma (Caco-2) and breast cancer (Mcf-7) cell lines.
The tris(hydroxymethyl) aminomethane is a simple organic compound commonly called TRIS with various other names including Trizma Base, or THAM and it is used widely as a buffering medium in many biochemical applications. The buffering pH range is 7 to 9, it can be used as a buffer and eluent in anion exchange chromatography. The metal ions and TRIS are often included together in biochemical studies[1, 2, 3, 4].
Bjerrum[4] has been discussed the extent to which one can make meaningful corrections for TRIS interaction with Mg++, Mn++, Fe++, Co++, Cu++, Zn++, Cd++, and Hg++ under conditions more appropriate to biochemical studies using a pH titration method[5]. Formation constants have been calculated, where possible from pH titration data. Hg++ and Cu++ interact strongly with TRIS, and Co++, Zn++, Cd++ much less[5]. The interaction of copper with TRIS has been studied by optical and ESR spectroscopic techniques. In solution, three different copper complexes are obtained as a function of pH. The Cu2+ ion is located on the two-fold axis and coordinated to the oxygen and nitrogen atoms of two TRIS moieties, these ligand atoms form the rectangular base of a pyramid structure in which oxygen from a water molecule acts as the fifth ligand[6].
TRIS is a part of the aminoalcohol group and has found many applications in biochemistry, medicine, biology and physiology[7]. It has also been reported that TRIS is used in the treatment of acidosis in acute lung injury and it is an effective method to control acidosis[8]. TRIS Schiff base derivatives are known to have a broad spectrum of biological activities including anti-tumour, antibiotic, anticancer, antihistamine, antifungal and anti-inflammatory effects. The crystal structures of some of the tris(hydroxymethyl)aminomethane Schiff base ligands have been reported by several research groups[9, 10, 11, 12].
In this study, a two iron(Ⅲ ) complexes containing TRIS ligand were prepared. The complexes were characterized by various spectroscopic techniques and further tested for their bacterial and cytotoxic activities.
All chemical reagents in this study are pure grade and were purchased from Sigma-Aldrich Chemical Company, USA. These chemicals are tris(hydroxymethyl) aminomethane (Fig.1), FeCl3 and Fe2(SO4)3. The analyses and the corresponding models are summarized as follows:
| Type of analysis | Models |
|---|---|
| Elemental analyses | Perkin Elmer CHN 2400 |
| Conductance | Jenway 4010 conductivity meter |
| FTIR spectra | Bruker FTIR Spectrophotometer |
| Electronic spectra | UV2 Unicam UV/Vis Spectrophotometer |
| Magnetic moment | Magnetic Susceptibility Balance |
| Thermo gravimetric | TG/DTG-50H, Shimadzu thermo-gravimetric analyzer |
| SEM | Quanta FEG 250 equipment |
| XRD | X ’ Pert PRO PAN analytical, with copper target |
| TEM | JEOL 100s microscopy |
The two iron(Ⅲ ) solid complexes were prepared by mixing 1.0 mmol of FeCl3 or Fe2(SO4)3 in 30 mL of distilled water with 2.0 mmol Tris (0.242 g) in 20 mL of methanol. The reaction mixtures were neutralized between pH=7~8 by 5% ammonia solution, then refluxed in a water bath for 2~3 h to give the precipitate. After cooling to room temperature, the solid complexes were filtered as fine precipitates. The precipitates were washed with hot distilled water and then methanol. Then they were dried and stored in a desiccators containing dry calcium chloride. The yield of the products was about 75%~80%. The complexes were subjected to elemental microanalyses for C, H, N, and iron ion contents and their structures were confirmed by their magnetic, molar conductance, thermo gravimetric, IR and electronic analyses. The solid products have a higher melting point -250 ℃. The micro analytical (calculated/Calc. and experimental/Found) and physical data of the two synthesized complexes are summarized in Table 1.
| Table 1 Micro analytical and physical data of two iron(Ⅲ ) Tris complexes |
Herein, a screening test for possible antibacterial activity was done using the two synthesized Tris compounds against four bacterial strains Klebsiella, Escherichia coli, Staphylococcus aureus, and Staphylococcus epidermidis. Using disc diffusion method[13] to test the antimicrobial effect. Inhibition zones diameters around the disc were determined.
In vitro the cytotoxic activity of the tested two iron(Ⅲ ) Tris complexes against two human cancer cell lines (colorectal adenocarcinoma (Caco-2) and breast cancer (MCF-7)) was evaluated using the standard neutral red uptake assay[14]. Cells from both cancer cell lines (from CURP) were grown in 25 cm3 cell culture flasks with complete DMEM medium which was supplemented with 10% Fetal bovine serum and 1% antibiotic (penicillin G potassium plus streptomycine) in humidified atmosphere and 5% CO2 at 37 ℃. Cells were grown until 80% confluent and the cultured monolayers were subjected to rinse twice with PBS. Cells were then trypsinized by 2 mL (0.25%) trypsin-EDTA solution for 1 min. To help cells detach from the flask surface, gentle agitation was done. Fresh complete DMEM medium was then added to stop the reaction. The cells suspension was then centrifuged at 1 600 r· min-1 for 5 min. the pellet was then re-suspended in 1 mL complete DMEM medium. Cell count was done using hemocytometer and the cell viability was checked using trypan blue (100% viability). A cell suspension was diluted in complete growth medium at 1× 105 cells· mL-1 approximately 2× 104 cells· well-1 was dispensed into 96 well plate for 24 hours. After cells were attached to the bottom surface of 96 well plate, the medium was gently discarded. All the six complexes were first dissolved in DMSO and then diluted in DMEM media. Different concentrations (10, 50 and 100 μ g· mL-1) from the tested complexes were prepared. 200 μ L from each treatment media was dispensed in 4 replicates for each concentration. Negative control was untreated cells with media only and the positive control was Doxorubicin HCL (8 μ g· mL-1). A 96 well plates was then incubated for 24 h at 37 ℃. Nutral red dye medium was also prepared and incubated at 37 ℃ for 24 h in order to remove the precipitated dye crystals centrifugation at 1 800 r· min-1 for 10 min was done. After the 24 hrs incubation period for the 96 well plate, medium was decanted from wells and 100 μ L of the prepared NR medium was added to each well and incubated again for 3 hrs at 37 ℃. The dye containing medium was decanted and each well was rinsed twice with 150 μ L PBS solution to remove excess dye remained in the wells. 150 μ L of destain solution was added and incubated for 10 min with shaking. The absorbance of solution was measured at 546 nm using microplate reader (BioTek, ELX808) and cell viability was calculated.
The two iron(Ⅲ ) Tris complexes were stable at room temperature and soluble in DMF and DMSO, but insoluble in ethanol, acetone, and benzene. The elemental analyses of the complexes (Table 1) showed that the iron ions form complexes with the composition NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and NH4[Fe(Tris)2(H2O)2] (Ⅱ ). The molar conductance of the complexes in DMSO at room temperature was 95 ohm-1· cm2· mol-1 for NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and 66 ohm-1· cm2· mol-1 for NH4[Fe(Tris)2(H2O)2] (Ⅱ ), indicating that the two comqlexes are in the range expected for 1:2 electrolytes[15]. The assumed structure of the complexes are presented in Fig.2.
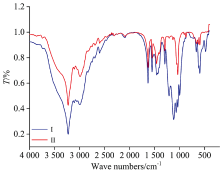
The infrared absorption spectra of the NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and NH4[Fe(Tris)2(H2O)2] (Ⅱ ) complexes were measured in the potassium bromide (Fig.3). Table 2 summarizes the characteristic infrared stretching frequencies for Tris ligand and the vibrational stretching frequencies for the two iron(Ⅲ ) complexes. The stretching frequencies of the hydroxyl groups in case of the free Tris ligand were expected to be in the 3 200~3 400 cm-1 region[16] and a strong broad band was observed around 3 290 cm-1 in the Tris ligand spectrum. However, no visible N— H stretching was observed, possibly due to the overlapping of the N— H stretching frequency with the O— H stretching frequency. The IR spectrum also evidenced the presence of
| Table 2 Infrared spectral data (cm-1) of iron(Ⅲ ) complexes |
The absorption bands of the free Tris ligand could be classified into two absorption regions of 200~260 and 310~350 nm due to n— π * and π — π * transitions, respectively. The electronic spectra of iron(Ⅲ ) complexes NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and NH4[Fe(Tris)2(H2O)2] (Ⅱ ) have been taken in solid state. The electronic spectra of the complexes exhibit three absorption bands at about 499~493, 375, and 330~336 nm, which may be assigned to transitions 6A1g→ 4T1g, 6A1g→ 4T2g, 6A1g→ 4A1g, 4Eg, respectively[20] and suggests octahedral geometry. The magnetic moment of the two complexes obtained in between 4.54~5.23 B.M. is good agreement for six-coordinated iron(Ⅲ ) system and consistent with the presence of a five-unpaired electrons[21].
The thermogravimetric (TGA/DrTGA) behaviors of the two synthesized iron(Ⅲ ) complexes Ⅰ and Ⅱ were studied under N2 atmosphere (Fig.4).
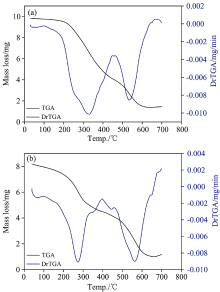 | Fig.4 TGA and DrTGA curves of (a): NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and (b): NH4[Fe(Tris)2(H2O)2] (Ⅱ ) |
The thermogravimetric analysis suggest that initially (first decomposition step) there is weight loss occurring in the 170~394 ℃ temperature range for two iron(Ⅲ ) complexes is attributed to as a loss of the water of Tris moieties, coordinated water molecules, and ammonia gas NH3. In the second step weight loss during 464~650 ℃ corresponding to remaining sulfate and Tris molecules, and remaining weight is in good agreement with iron(Ⅲ ) oxide (FeO) contaminated with few carbon atoms.
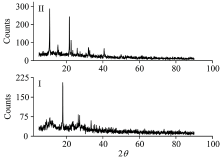
X-ray powder diffraction patterns of NH4[Fe2(Tris)2(H2O)4(SO4)] (Ⅰ ) and NH4[Fe(Tris)2(H2O)2] (Ⅱ ) complexes within 2θ =5~90° are displayed in Fig.5, these patterns have semi-crystalline nature. The size of particles was estimated based on the full-width at half-maximum (FWHM) using Scherrer relationship[22]. The distinguish XRD peaks of the iron(Ⅲ ) complexes I and Ⅱ are detected at 2θ (° )=(9.12, 10.96, 18.04, 23.28, 26.50, 27.40 and 33.58) and (10.95, 15.30, 21.69, 22.61, 26.04, 27.40, 30.59, 32.20 and 40.66), respectively due to iron metal ion and Tris ligand. The TEM images of the synthesized iron(Ⅲ ) complexes of Ⅰ and Ⅱ are shown in Fig.6, the complexes refer to a spherical nanoparticles within 100~200 nm and 7~16 nm, respectively. SEM images of both iron(Ⅲ ) complexes (Fig.6) were scanned at energy within 10-30 kV with magnification × 20 000~40 000. From Fig.6 it is clear that the morphology of the synthesized iron(Ⅲ ) complexes have a flower-shaped structure with the aggregation of tremendous micro-size fine powders and homogeneous uniform.
The antibacterial effect of the two iron(Ⅲ ) synthesized complexes in this study was assessed by screening test using disc diffusion method. Four bacterial strains such as Klebsiella (G-), Escherichia coli (G-), Staphylococcus aureus (G+) and Staphylococcus epidermidis (G+) were used in the investigation. The results showed no any antibacterial activity for complexes Ⅰ and Ⅱ on Escherichia coli, Staphylococcus aureus and Staphylococcus epidermidis. However, weak antibacterial effect was seen with both iron(Ⅲ ) complexes against Klebsiella only, with inhibition zones of 0.1 and 0.2 mm respectively, as compared with the standard antibiotic gentamycin with inhibition zone of 0.3 mm.
In this study, the anticancer effect of the two iron(Ⅲ ) complexes was investigated by using neutral red uptake assay on two human cancer cell lines. Colorectal adenocarcinoma cell line (Caco-2) and breast cancer cell line (MCF-7) were used to test the cytotoxicity of complexes Ⅰ and Ⅱ . Complex Ⅰ showed low cytotoxic activity with IC50 value > 100 μ g· mL-1 in Colorectal adenocarcinoma cell line (Caco-2), however complex Ⅰ has no any cytotoxicity on breast cancer cell line (MCF-7) at lower and higher concentration. Complex Ⅱ showed no cytotoxic effect on (Caco-2) cell line at lower concentration.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|